Development of SNP parentage assignment techniques in the yellowfin seabream Acanthopagrus latus
-
Abstract: Acanthopagrus latus is an essential aquaculture species on the south coast of China. However, there is a lack of systematic breeding of A. latus, which considerably limits the sustainable development of A. latus. As a result, genetic improvements are urgently needed to breed new strains of A. latus with rapid growth and strong resistance to disease. During selective breeding, it is necessary to estimate the genetic parameters of the target trait, which in turn depends on an accurate disentangled pedigree for the selective population. Therefore, it is necessary to establish the parentage assignment technique for A. latus. In this study, 95 individuals selected from their parents and their 14 families were used as experimental material. SNPs were developed by genome re-sequencing, and highly polymorphic SNPs were screened on the basis of optimized filtering parameters. A total of 14 392 738 SNPs were discovered and 205 SNPs were selected for parentage assignment using the CERVUS software. In the model where the gender of the parents is known, the assignment success rate is 98.61% for the male parent, 97.22% for the female parent, and 95.83% for the parent pair. In the model where the gender of the parents is unknown, the assignment success rate is 100% for a single parent and 90.28% for the parent pair. The results of this study were expected to serve as a reference for the breeding of new varieties of A.latus.
-
Key words:
- Acanthopagrus latus /
- parentage assignment /
- SNP /
- Genome re-sequencing
-
Table 1. Parental combinations and number of samples per family
Family Id Female parent Male parent Number of offspring taken J01 d01 s01 4 J05 d05 s02 5 J06 d06 s02 5 J09 d09 s03 3 J10 d10 s04 6 J11 d11 s04 4 J17 d17 s07 4 J18 d18 s07 7 J19 d19 s07 5 J20 d20 s08 6 J23 d23 s09 6 J25 d25 s09 5 J26 d26 s10 6 J30 d30 s11 6 Total 14 9 72 Table 2. SNP annotation information statistics in A. latus
Category Number of SNP Proportion of total /% Exonic 579 057 4.02 Exonic Stop gain 2 310 0.02 Exonic Stop loss 247 0.00 Exonic Synonymous 337 298 2.34 Exonic Non-synonymous 239 202 1.66 Intronic 7 161 298 49.76 Splicing 1 915 0.01 Upstream 349 101 2.43 Downstream 342 049 2.38 Upstream/Downstream 82 478 0.57 Intergenic 3 836 766 26.66 Others 2 040 243 14.18 Total 14 392 738 − Table 3. Parentage assignment results of five sets of SNPs (with 95 % confidence)
Number
of SNPModel of known parental gender Model of unknown parental gender Assignment success
rates of male parentAssignment success
rates of female parentAssignment success
rates of parent pairAssignment success
rates of single parentAssignment success
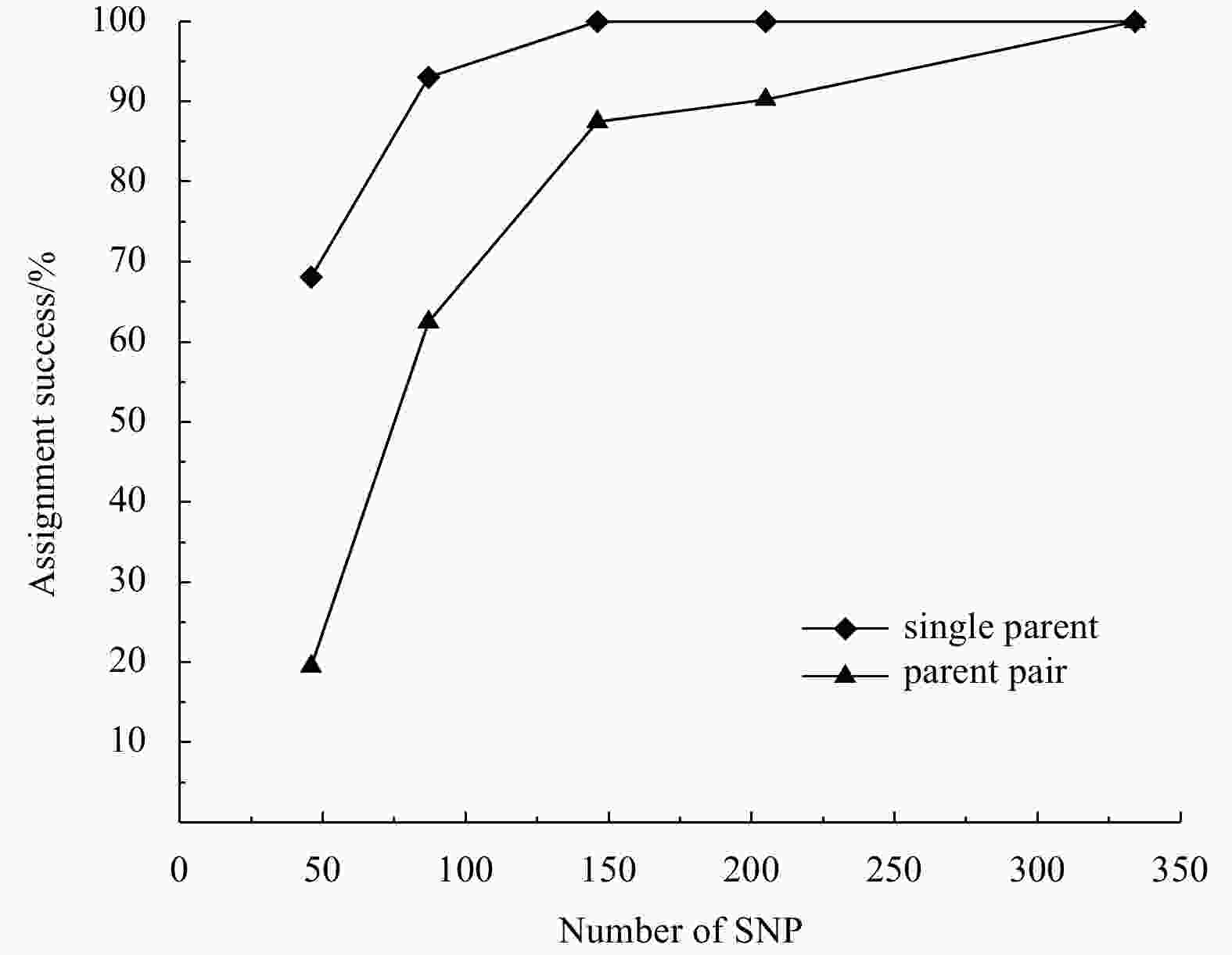
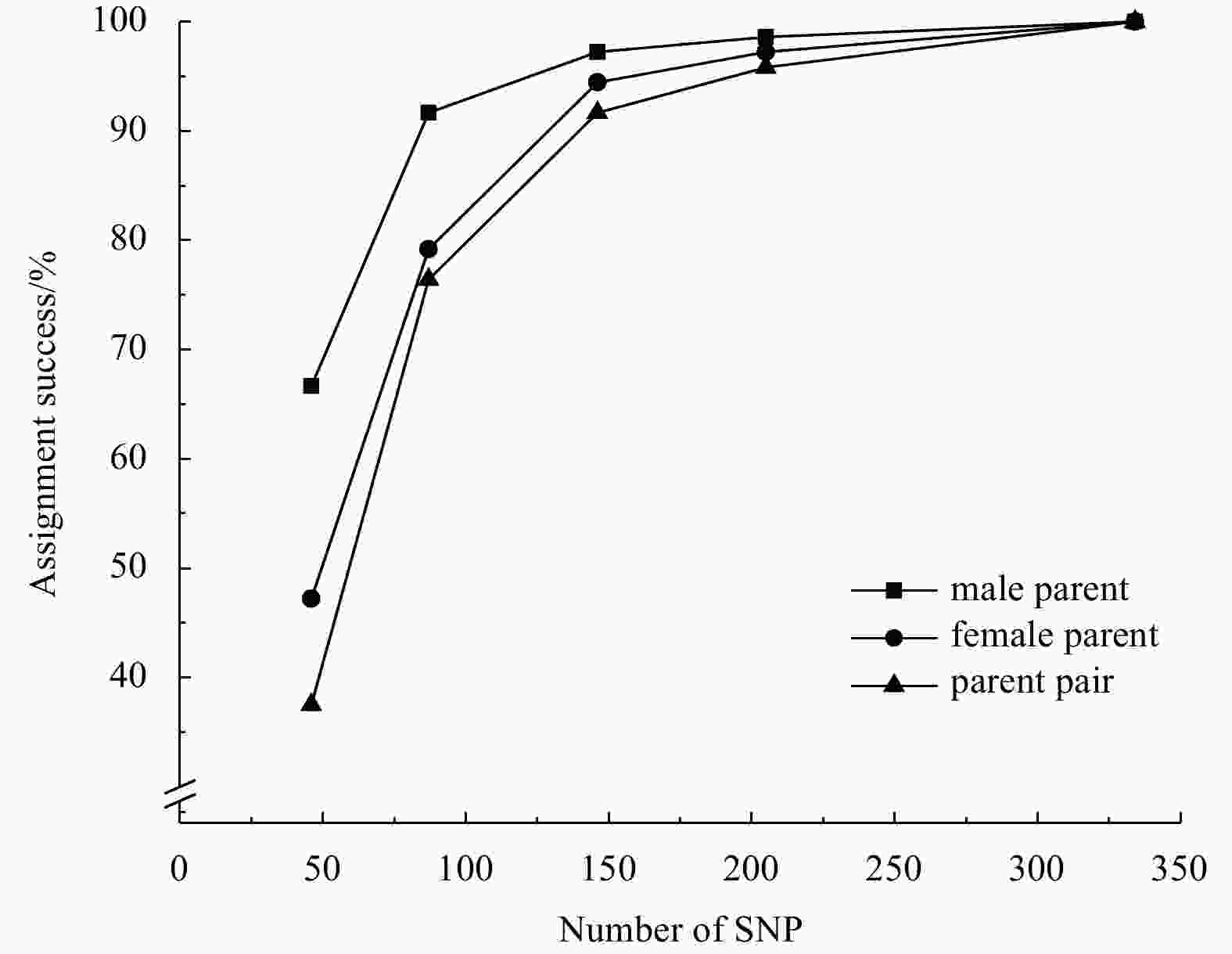
rates of parent pair46 66.67% 47.22% 37.50% 68.06% 19.44% 87 91.67% 79.17% 76.39% 93.06% 62.50% 146 97.22% 94.44% 91.67% 100% 87.50% 205 98.61% 97.22% 95.83% 100% 90.28% 334 100% 100% 100% 100% 100% Notes: Grey shading indicates the best combination of markers. -
Abadía-Cardoso A, Anderson E C, Pearse D E, et al. 2013. Large-scale parentage analysis reveals reproductive patterns and heritability of spawn timing in a hatchery population of steelhead ( Oncorhynchus mykiss). Molecular Ecology, 22(18): 4733–4746, doi: 10.1111/mec.12426 Anderson E C, Garza J C. 2006. The power of single-nucleotide polymorphisms for large-scale parentage inference. Genetics, 172(4): 2567–2582, doi: 10.1534/genetics.105.048074 Flanagan S P, Jones A G. 2019. The future of parentage analysis: From microsatellites to SNPs and beyond. Molecular Ecology, 28(3): 544–567, doi: 10.1111/mec.14988 García-Fernández C, Sánchez J A, Blanco G. 2018. SNP-haplotypes: An accurate approach for parentage and relatedness inference in gilthead sea bream ( Sparus aurata). Aquaculture, 495: 582–591, doi: 10.1016/j.aquaculture.2018.06.019 Gjedrem T, Robinson N, Rye M. 2012. The importance of selective breeding in aquaculture to meet future demands for animal protein: a review. Aquaculture, 350–353: 117–129, doi: 10.1016/j.aquaculture.2012.04.008 Harlizius B, Lopes M S, Duijvesteijn N, et al. 2011. A single nucleotide polymorphism set for paternal identification to reduce the costs of trait recording in commercial pig breeding. Journal of Animal Science, 89(6): 1661–1668, doi: 10.2527/jas.2010-3347 Hauser L, Baird M, Hilborn R, et al. 2011. An empirical comparison of SNPs and microsatellites for parentage and kinship assignment in a wild sockeye salmon ( Oncorhynchus nerka) population. Molecular Ecology Resources, 11(S1): 150–161, doi: 10.1111/j.1755-0998.2010.02961.x Holman L E, De La Serrana D G, Onoufriou A, et al. 2017. A workflow used to design low density SNP panels for parentage assignment and traceability in aquaculture species and its validation in Atlantic salmon. Aquaculture, 476: 59–64, doi: 10.1016/j.aquaculture.2017.04.001 Houston R D, Bean T P, Macqueen D J, et al. 2020. Harnessing genomics to fast-track genetic improvement in aquaculture. Nature Reviews Genetics, 21(7): 389–409, doi: 10.1038/s41576-020-0227-y Kalinowski S T, Taper M L, Marshall T C. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology, 16(5): 1099–1106, doi: 10.1111/j.1365-294X.2007.03089.x Li Heng, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25(14): 1754–1760, doi: 10.1093/bioinformatics/btp324 Liu Sixin, Palti Y, Gao Guangtu, et al. 2016. Development and validation of a SNP panel for parentage assignment in rainbow trout. Aquaculture, 452: 178–182, doi: 10.1016/j.aquaculture.2015.11.001 Marshall T C, Slate J, Kruuk L E B, et al. 1998. Statistical confidence for likelihood-based paternity inference in natural populations. Molecular Ecology, 7(5): 639–655, doi: 10.1046/j.1365-294x.1998.00374.x Slate J, Marshall T, Pemberton J. 2000. A retrospective assessment of the accuracy of the paternity inference program CERVUS. Molecular Ecology, 9(6): 801–808, doi: 10.1046/j.1365-294x.2000.00930.x Steele C A, Anderson E C, Ackerman M W, et al. 2013. A validation of parentage-based tagging using hatchery steelhead in the Snake River basin. Canadian Journal of Fisheries and Aquatic Sciences, 70(7): 1046–1054, doi: 10.1139/cjfas-2012-0451 Tokarska M, Marshall T, Kowalczyk R, et al. 2009. Effectiveness of microsatellite and SNP markers for parentage and identity analysis in species with low genetic diversity: the case of European bison. Heredity, 103(4): 326–332, doi: 10.1038/hdy.2009.73 Vandeputte M, Haffray P. 2014. Parentage assignment with genomic markers: a major advance for understanding and exploiting genetic variation of quantitative traits in farmed aquatic animals. Frontiers in Genetics, 5: 432, doi: 10.3389/fgene.2014.00432 Walling C A, Pemberton J M, Hadfield J D, et al. 2010. Comparing parentage inference software: reanalysis of a red deer pedigree. Molecular Ecology, 19(9): 1914–1928, doi: 10.1111/j.1365-294X.2010.04604.x Weinman L R, Solomon J W, Rubenstein D R. 2015. A comparison of single nucleotide polymorphism and microsatellite markers for analysis of parentage and kinship in a cooperatively breeding bird. Molecular Ecology Resources, 15(3): 502–511, doi: 10.1111/1755-0998.12330 Xu Jian, Feng Jingyan, Peng Wenzhu, et al. 2017. Development and evaluation of a high-throughput single nucleotide polymorphism multiplex assay for assigning pedigrees in common carp. Aquaculture Research, 48(4): 1866–1876, doi: 10.1111/are.13024 Yue Genhua, Xia Junhong. 2014. Practical considerations of molecular parentage analysis in fish. Journal of the World Aquaculture Society, 45(2): 89–103, doi: 10.1111/jwas.12107 Zhao Honggang, Li Chao, Hargrove J S, et al. 2018. SNP marker panels for parentage assignment and traceability in the Florida bass ( Micropterus floridanus). Aquaculture, 485: 30–38, doi: 10.1016/j.aquaculture.2017.11.014 Zheng Guobin, Zhao Hongbo, Huang Liangmin, et al. 2023. Discovery and verification of SNP in Acanthopagrus latus. Journal of Tropical Oceanography (in Chinese), 42(2): 78–86, doi: 10.11978/2022108 Zhu Kecheng, Song Ling, Liu Baosuo, et al. 2020. Establishment of parentage determination in yellowfin seabream ( Acanthopagrus latus). Journal of Fisheries of China (in Chinese), 44(3): 351–357, doi: 10.11964/jfc.20181011480 -




 下载:
下载: