Diversity of protease-producing bacteria in the Bohai Bay sediment and their extracellular enzymatic properties
-
Abstract: Protease-producing bacteria play key roles in the degradation of organic nitrogen materials in marine sediments. However, their diversity, production of proteases and other extracellular enzymes, even in situ ecological functions remain largely unknown. In this study, we investigated the diversity of cultivable extracellular protease-producing bacteria in the sediments of the Bohai Bay. A total of 109 bacterial isolates were obtained from the sediments of 7 stations. The abundance of cultivable protease-producing bacteria was about 104 CFU/g of sediment in all the samples. Phylogenetic analysis based on 16S rRNA gene sequences classified all the isolates into 14 genera from phyla Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria, with Pseudoalteromonas (63/109, 57.8%), Bacillus (9/109, 8.2%), Sulfitobacter (8/109, 7.3%) and Salegentibacter (6/109, 5.5%) as the dominant taxa. Enzymatic inhibition tests indicated that all the tested isolates produced serine and/or metalloprotease, with only a small proportion producing cysteine and/or aspartic proteases. Several extracellular enzyme activities, including alginase, lipase, amylase and cellulose, and nitrate reduction were also detected for strains with higher protease activities. According the results, the protease-producing bacteria could also be participate in many biogeochemical processes in marine sediments. Our study broadened understanding and knowledge on the potential ecological functions of protease-producing bacteria in marine sediments.
-
Key words:
- protease-producing bacteria /
- diversity /
- inhibition test /
- multi-extracellular enzymes
-
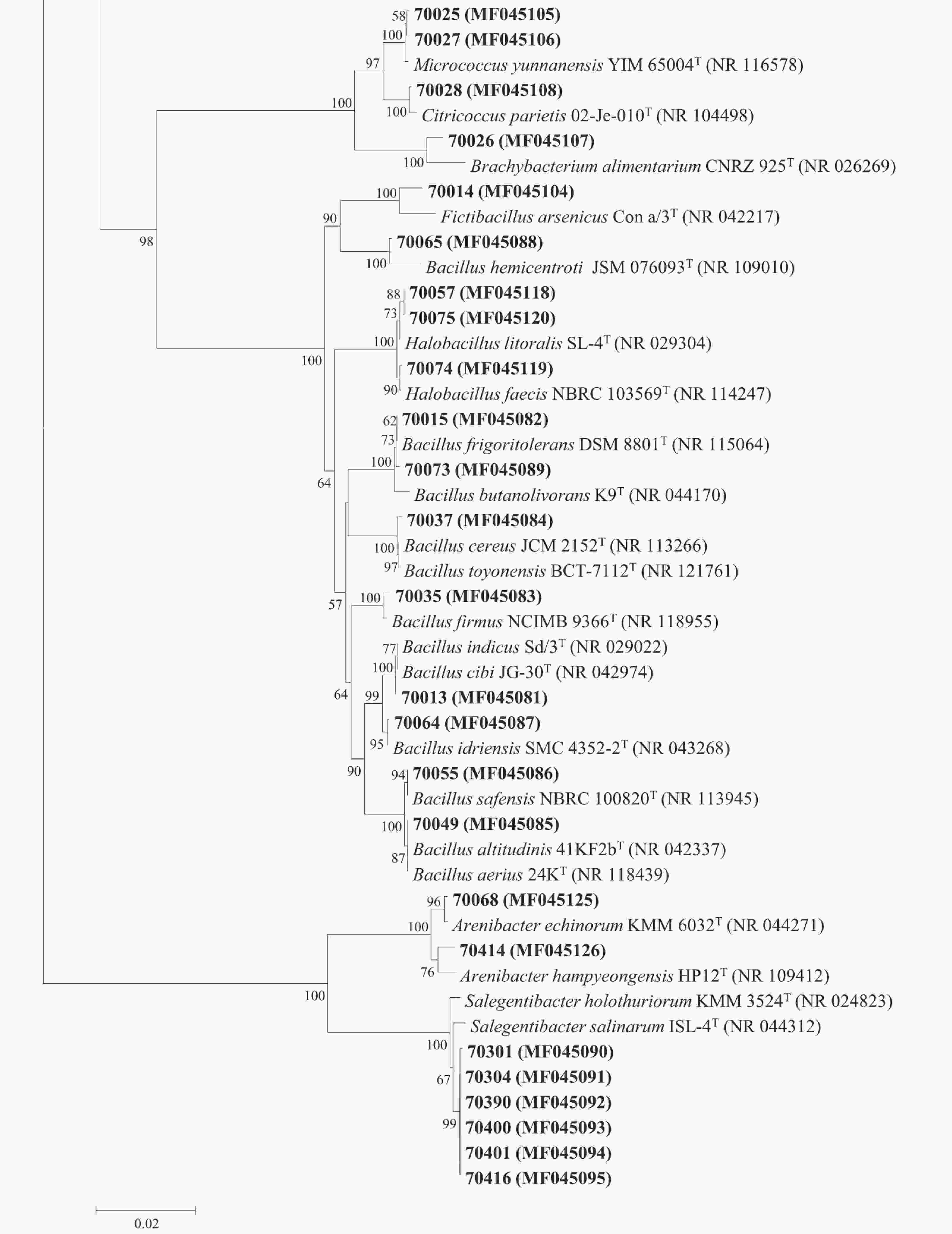
Figure 2. Phylogenetic tree of the protease-producing bacteria isolated from the Bohai Bay based on 16S rRNA gene sequences. Taxa and GenBank accession numbers in boldface were generated in this study. The tree was constructed by neighbor-joining method using MEGA version 6.0. Only bootstrap values greater than 50% are presented in the nodes. The scale bar represents 2% nucleotide substitution.
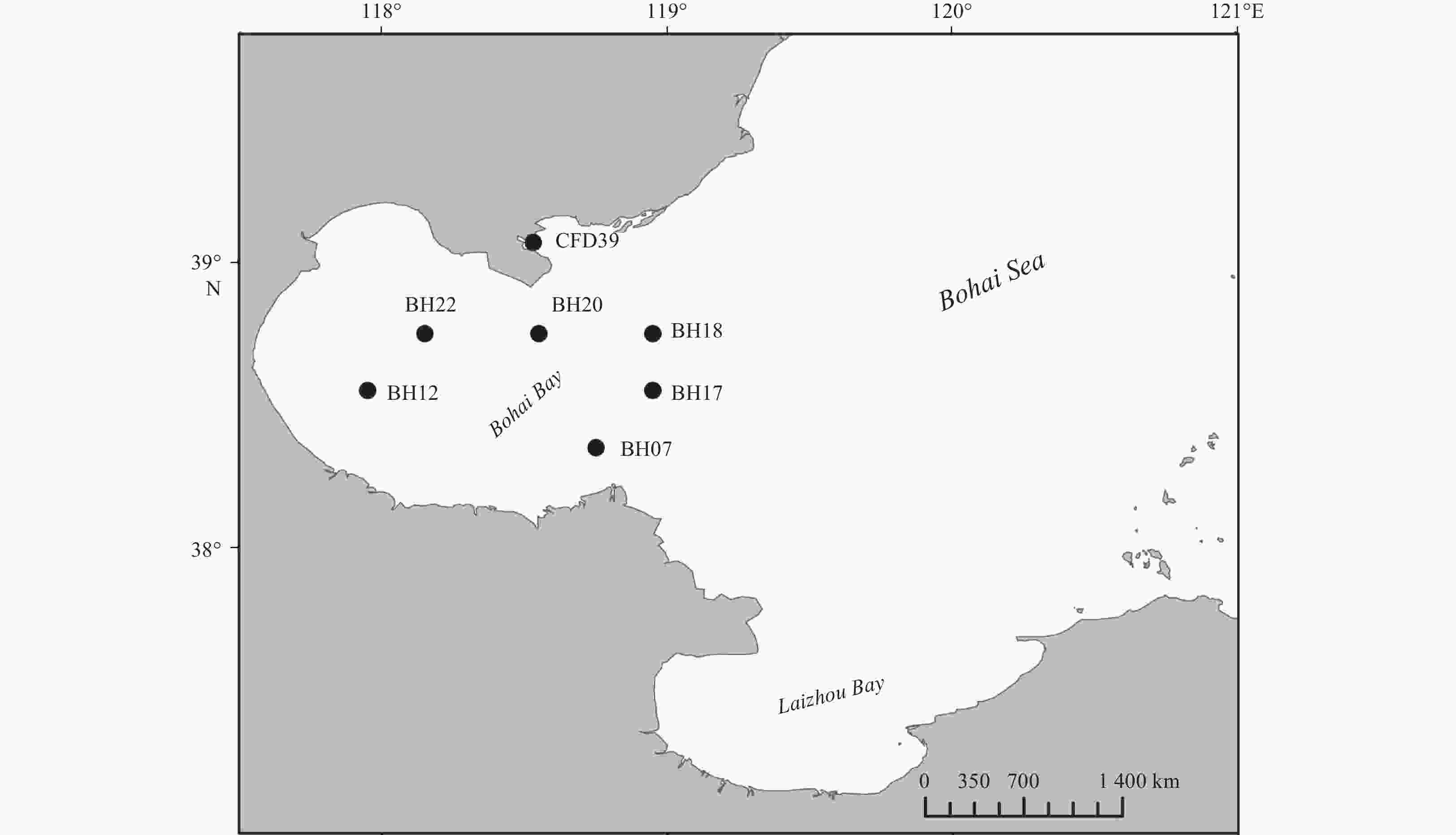
Table 1. Characteristics of the sampling stations and the distribution of different genera in these stations
Properties Station BH07 BH12 BH17 BH18 BH20 BH22 CFD39 GPS 38.35°N, 118.75°E 38.55°N, 117.95°E 38.55°N, 118.95°E 38.75°N, 118.95°E 38.75°N, 118.55°E 38.75°N, 118.15°E 39.07°N, 118.53°E Characteristics for sediment samples Depth/m 16.14 9.66 22.65 25.57 23.78 17.27 12.19 Temperature/°C 24.97 27.11 20.36 19.32 23.27 25.47 26.14 pH 8.11 8.08 7.91 7.90 8.01 8.18 8.08 DO 4.02 4.17 2.20 2.28 3.50 5.11 4.86 Sal 30.30 30.02 30.31 30.45 30.51 30.20 30.63 OrgC/% 0.42 0.51 0.55 0.60 0.54 0.57 0.87 OrgN/% 0.06 0.08 0.09 0.08 0.09 0.10 0.11 C/N1) 7.00 6.38 6.11 7.5 6.00 5.70 7.91 Genera distribution Proteobacteria Pseudoalteromonas 3 8 15 6 16 7 8 Shewanella 3 1 Marinobacter 1 3 Sulfitobacter 4 4 Celeribacter 1 1 1 Albirhodobacter 1 1 Firmicutes Bacillus 7 1 1 Halobacillus 1 1 1 Fictibacillus 1 Actinobacteria Micrococcus 1 1 Citricoccus 1 Brachybacterium 1 Bacteroidetes Arenibacter 2 Salegentibacter 2 2 2 Total strain number (109) Diversity index Shannon–Wiener (H′) 1.58 1.22 1.66 1.01 0.34 0.94 0.64 Simpson (D) 0.72 0.61 0.71 0.61 0.20 0.48 0.34 Pielou (J) 0.81 0.76 0.76 0.92 0.50 0.68 0.58 Note: 1) C/N is the abbreviation of OrgC/OrgN. Table 2. Summary of the inhibition test and the extracellular enzyme production analyses of the tested strains isolated from the Bohai Bay
Genera Strain Inhibition ratio1) (I)/% H/C ratio2) Nitrate PMSF O-P E-64 P-A Casein Gelatin Elastin Amylase Cellulase Alginase Tween 80 Pseudoalteromonas 70004 43.64 22.87 0 0 1.77 3.00 1.27 0 0 0 0 – 70006 60.97 0 3.35 0 2.78 2.92 1.40 4.00 2.18 3.26 6.31 + 70032 80.75 46.83 0 0 1.54 3.10 0 0 0 0 1.70 70038 16.89 0 0 0 2.33 4.50 0 0 0 0 1.61 – 70039 43.83 0 0 0 2.50 4.71 0 0 3.68 1.11 3.27 70048 48.41 16.53 0 0 2.78 5.00 0 0 0 0 4.52 + 70051 71.35 45.19 0 2.99 3.25 2.67 1.86 0 0 0 2.59 – 70053 76.43 29.47 0 0 2.25 5.00 0 0 0 1.49 5.40 – 70056 80.05 0 0 0 2.78 4.14 0 0 0 3.60 0 – 70305 46.37 0 0 0 3.42 1.40 2.21 1.59 0 1.20 3.17 – 70306 49.18 34.79 4.11 0 2.87 3.22 1.58 1.76 1.08 1.25 1.51 + 70307 81.10 44.84 0 0 2.94 2.56 1.92 3.23 0 1.33 3.36 – 70308 99.23 48.20 5.80 0 2.75 2.87 1.58 2.07 0 1.16 3.83 + 70320 52.13 5.81 0 0 2.31 1.46 1.55 1.49 1.22 1.22 1.51 – 70325 39.57 6.21 0 0 3.14 1.27 1.70 1.98 0 1.44 2.99 + 70326 81.42 34.55 0 0 3.19 1.14 2.10 2.19 0 1.96 1.70 – 70333 29.63 6.34 0 0 2.62 1.20 1.55 1.64 0 1.20 2.79 – 70339 90.13 31.28 4.48 4.48 2.00 0 1.50 1.61 4.11 1.23 3.46 + 70342 85.92 3.20 0 0 2.18 1.45 1.58 0 0 0 0 – 70344 70.04 53.84 7.04 0 2.38 1.42 1.25 1.34 0 1.28 2.96 – 70354 51.04 20.35 4.25 0 3.43 2.85 1.73 1.26 0 1.32 1.33 – 70356 68.40 10.60 0 7.72 3.18 1.57 2.09 1.75 0 1.31 1.19 – 70359 77.04 42.95 0 3.74 3.42 1.80 1.60 1.83 0 1.86 2.64 + 70361 50.74 6.17 3.60 0 3.00 2.36 1.77 1.35 0 2.12 2.83 – 70365 92.29 27.77 0 0 3.21 3.29 2.50 1.34 0 2.18 0 + 70369 57.37 34.39 0 0 2.44 2.08 1.50 1.76 1.41 2.37 1.18 – 70373 95.19 44.79 0 3.12 3.29 3.17 1.50 2.97 1.12 3.36 2.73 + 70374 76.82 37.19 0 0 3.13 2.27 1.36 2.36 0 6.55 2.78 – 70375 61.08 0 2.23 0 2.93 2.88 2.11 2.89 0 4.63 0 – 70376 15.39 5.92 0 0 3.00 2.75 1.33 0 0 2.16 0 – 70379 42.46 51.17 3.70 0 2.80 2.64 1.80 1.27 0 3.29 2.79 – 70380 91.52 51.24 1.28 4.57 2.82 3.15 1.64 0 1.59 2.10 0 – 70413 99.18 32.76 0 0 2.29 1.27 1.33 1.40 0 2.62 2.71 – Bacillus 70013 63.69 8.67 0 0 1.80 3.67 0 0 0 8.74 0 – 70035 9.42 0 0 0 1.40 6.50 0 0 0 0 0 + 70037 19.80 3.89 0 0 2.14 7.00 0 0 0 2.96 2.04 – 70049 92.58 44.74 0 3.09 4.80 5.42 1.32 0 0 0 0 – 70055 31.98 18.38 7.45 7.63 1.36 3.86 0 0 0 0 0 – Fictibacillus 70014 43.21 1.84 0 0 1.67 5.00 0 0 0 2.52 0 – Micrococcus 70025 68.60 24.30 7.83 0 4.00 5.17 0 0 0 0 0 – 70027 61.26 17.12 9.01 0 3.80 4.14 0 0 0 1.88 0 – Halobacillus 70074 38.71 0 0 0 3.57 3.33 2.29 0 0 2.95 0 – 70075 63.57 15.94 0 0 2.00 4.00 0 0 1.37 0 0 – Salegentibacter 70304 35.37 6.19 7.24 0 4.60 1.29 5.00 0 0 2.63 0 + 70390 84.24 0 0 0 8.33 7.83 5.40 0 0 3.41 0 + 70400 42.39 0 0 0 5.00 4.60 3.57 0 0 2.00 0 – 70401 46.26 8.49 0 0 6.75 6.00 5.00 0 0 2.62 0 + 70416 20.22 1.31 0 0 6.00 3.00 5.50 0 0 2.11 0 + Marinobacter 70434 41.70 37.85 0 0 2.09 0 0 3.54 0 5.27 2.40 + 70435 38.74 35.58 0 0 1.28 3.19 0 3.82 0 7.84 2.40 + Note: 1) Inhibition ratio (I, %) was calculated by using control activity minus the relative activity of a sample with an inhibitor and the activity of a sample without any inhibitor was taken as a control. 2) H/C ratio is the ratio of the hydrolytic zone diameter versus the colony diameter of a colony on the plate. PMSF, phenylmethylsulfonyl fluoride; OP, 1, 10-phenanthroline; P-A, pepstatin A. -
Akagawa-Matsushita M, Matsuo M, Koga Y, et al. 1993. Alteromonas atlantica sp. nov. and Alteromonas carrageenovora sp. nov., bacteria that decompose algal polysaccharides. International Journal of Systematic and Bacteriology, 43(2): 400–400. doi: 10.1099/00207713-43-2-400 Alasalvar C, Taylor K D A, Zubcov E, et al. 2002. Differentiation of cultured and wild sea bass (Dicentrarchus labrax): Total lipid content, fatty acid and trace mineral composition. Food Chemistry, 79(2): 145–150. doi: 10.1016/S0308-8146(02)00122-X Anastasakis K, Ross A B, Jones J M. 2011. Pyrolysis behaviour of the main carbohydrates of brown macro-algae. Fuel, 90(2): 598–607. doi: 10.1016/j.fuel.2010.09.023 Bowman J P. 2007. Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Marine Drugs, 5(4): 220–241. doi: 10.3390/md504220 Chen Xiulan, Zhang Yuzhong, Gao Peiji, et al. 2003. Two different proteases produced by a deep-sea psychrotrophic bacterial strain, Pseudoaltermonas sp. SM9913. Marine Biology, 143(5): 989–993. doi: 10.1007/s00227-003-1128-2 Chi W J, Park J S, Kang D K, et al. 2014. Production and characterization of a novel thermostable extracellular agarase from Pseudoalteromonas hodoensis newly isolated from the West Sea of South Korea. Applied Biochemistry and Biotechnology, 173(7): 1703–1716. doi: 10.1007/s12010-014-0958-3 Dang Hongyue, Zhu Hu, Wang Jing, et al. 2009. Extracellular hydrolytic enzyme screening of culturable heterotrophic bacteria from deep-sea sediments of the Southern Okinawa Trough. World Journal of Microbiology and Biotechnology, 25(1): 71–79. doi: 10.1007/s11274-008-9865-5 Demirbas A, DemiRbas M F. 2011. Importance of algae oil as a source of biodiesel. Energy Conversion and Management, 52(1): 163–170. doi: 10.1016/j.enconman.2010.06.055 Engel A S, Porter M L, Stern L A, et al. 2004. Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria”. FEMS Microbiology Ecology, 51(1): 31–53. doi: 10.1016/j.femsec.2004.07.004 Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution, 39(4): 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x Feng Huan, Jiang Hongyou, Gao Wensheng, et al. 2011. Metal contamination in sediments of the western Bohai Bay and adjacent estuaries, China. Journal of Environmental Management, 92(4): 1185–1197. doi: 10.1016/j.jenvman.2010.11.020 Fu Yanzhao, Xu Shiguo, Liu Jianwei. 2016. Temporal-spatial variations and developing trends of Chlorophyll-a in the Bohai Sea, China. Estuarine, Coastal and Shelf Science, 173: 49–56. doi: 10.1016/j.ecss.2016.02.016 Gamal R F, El-Tayeb T S, Raffat E I, et al. 2016. Optimization of chitin yield from shrimp shell waste by Bacillus subtilis and impact of gamma irradiation on production of low molecular weight chitosan. International Journal of Biological Macromolecules, 91: 598–608. doi: 10.1016/j.ijbiomac.2016.06.008 Hansen G H, Sørheim R. 1991. Improved method for phenotypical characterization of marine bacteria. Journal of Microbiological Methods, 13(3): 231–241. doi: 10.1016/0167-7012(91)90049-V He Peiqing, Li Li, Liu Jihua, et al. 2016. Diversity and distribution of catechol 2, 3-dioxygenase genes in surface sediments of the Bohai Sea. FEMS Microbiology Letters, 363(10): fnw086. doi: 10.1093/femsle/fnw086 Hill T C J, Walsh K A, Harris J A, et al. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiology Ecology, 43(1): 1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x Hu Ningjing, Shi Xuefa, Liu Jihua, et al. 2010. Concentrations and possible sources of PAHs in sediments from Bohai Bay and adjacent shelf. Environmental Earth Sciences, 60(8): 1771–1782. doi: 10.1007/s12665-009-0313-0 Hunter E M, Mills H J, Kostka J E. 2006. Microbial community diversity associated with carbon and nitrogen cycling in permeable shelf sediments. Applied and Environmental Microbiology, 72(9): 5689–5701. doi: 10.1128/AEM.03007-05 Itoh T. 1990. Cellulose synthesizing complexes in some giant marine algae. Journal of Cell Science, 95(2): 309–319 Kitamikado M, Yamaguchi K, Tseng C H, et al. 1990. Method designed to detect alginate-degrading bacteria. Applied and Environmental Microbiology, 56(9): 2939–2940. doi: 10.1128/AEM.56.9.2939-2940.1990 Li Yan, Wu Chaoya, Zhou Mingyang, et al. 2017. Diversity of cultivable protease-producing bacteria in Laizhou Bay sediments, Bohai Sea, China. Frontiers in Microbiology, 8: 405. doi: 10.3389/fmicb.2017.00405 Mondol M M A M, Shin H J, Islam M T. 2013. Diversity of secondary metabolites from marine Bacillus species: Chemistry and biological activity. Marine Drugs, 11(8): 2846–2872. doi: 10.3390/md11082846 Mu Di, Yuan Dekui, Feng Huan, et al. 2017. Nutrient fluxes across sediment-water interface in Bohai Bay Coastal Zone, China. Marine Pollution Bulletin, 114(2): 705–714. doi: 10.1016/j.marpolbul.2016.10.056 Olivera N L, Sequeiros C, Nievas M L. 2007. Diversity and enzyme properties of protease-producing bacteria isolated from sub-Antarctic sediments of Isla de Los Estados, Argentina. Extremophiles, 11(3): 517–526. doi: 10.1007/s00792-007-0064-3 Pujalte M J, Sitjà-Bobadilla A, Macián M C, et al. 2007. Occurrence and virulence of Pseudoalteromonas spp. in cultured gilthead sea bream (Sparus aurata L.) and European sea bass (Dicentrarchus labrax L.). Molecular and phenotypic characterisation of P. undina strain U58. Aquaculture, 271(1–4): 47–53. doi: 10.1016/j.aquaculture.2007.06.015 Putz M, Schleusner P, Rütting T, et al. 2018. Relative abundance of denitrifying and DNRA bacteria and their activity determine nitrogen retention or loss in agricultural soil. Soil Biology and Biochemistry, 123: 97–104. doi: 10.1016/j.soilbio.2018.05.006 R Core Team. 2014. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing, 2013 Saitou N, Nei M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4(4): 406–425. doi: 10.1093/oxfordjournals.molbev.a040454 Sanger F, Nicklen S, Coulson A R. 1977. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences of the United States of America, 74(12): 5463–5467. doi: 10.1073/pnas.74.12.5463 Sfanos K, Harmody D, Dang P, et al. 2005. A molecular systematic survey of cultured microbial associates of deep-water marine invertebrates. Systematic and Applied Microbiology, 28(3): 242–264. doi: 10.1016/j.syapm.2004.12.002 Smant G, Stokkermans J P W G, Yan Yitang, et al. 1998. Endogenous cellulases in animals: Isolation of β-1, 4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proceedings of the National Academy of Sciences of the United States of America, 95(9): 4906–4911. doi: 10.1073/pnas.95.9.4906 Sun Jinsheng, Guo Fei, Geng Xuyun, et al. 2011. Seasonal changes and diversity of bacteria in Bohai Bay by RFLP analysis of PCR-amplified 16S rDNA gene fragments. World Journal of Microbiology and Biotechnology, 27(2): 275–284. doi: 10.1007/s11274-010-0456-x Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12): 2725–2729. doi: 10.1093/molbev/mst197 Thamdrup B, Dalsgaard T. 2008. Nitrogen cycling in sediments. In: Kirchman D L, ed. Microbial Ecology of the Oceans. 2nd ed. Hoboken, NJ: John Wiley & Sons, Inc., 527–568 Tsekos I. 1999. The sites of cellulose synthesis in algae: Diversity and evolution of cellulose-synthesizing enzyme complexes. Journal of Phycology, 35(4): 635–655. doi: 10.1046/j.1529-8817.1999.3540635.x Viola R, Nyvall P, Pedersén M. 2001. The unique features of starch metabolism in red algae. Proceedings of the Royal Society B: Biological Sciences, 268(1474): 1417–1422. doi: 10.1098/rspb.2001.1644 Yadav A N, Sachan S G, Verma P, et al. 2015. Cold active hydrolytic enzymes production by psychrotrophic Bacilli isolated from three sub-glacial lakes of NW Indian Himalayas. Journal of Basic Microbiology, 56(3): 294–307. doi: 10.1002/jobm.201500230 Zhang Xiying, Han Xiaoxu, Chen Xulan, et al. 2015. Diversity of cultivable protease-producing bacteria in sediments of Jiaozhou Bay, China. Frontiers in Microbiology, 6: 1021. doi: 10.3389/fmicb.2015.01021 Zhang Quansheng, Tang Xuexi, Cong Yizhou, et al. 2007. Breeding of an elite Laminaria variety 90–1 through inter-specific gametophyte crossing. Journal of Applied Phycology, 19(4): 303–311. doi: 10.1007/s10811-006-9137-4 Zhao Huilin, Chen Xiulan, Xie Binbin, et al. 2012. Elastolytic mechanism of a novel M23 metalloprotease pseudoalterin from deep-sea Pseudoalteromonas sp. CF6–2: Cleaving not only glycyl bonds in the hydrophobic regions but also peptide bonds in the hydrophilic regions involved in cross-linking. Journal of Biological and Chemistry, 287(47): 39710–39720. doi: 10.1074/jbc.M112.405076 Zhou Mingyang, Chen Xiulan, Zhao Huilin, et al. 2009. Diversity of both the cultivable protease-producing bacteria and their extracellular proteases in the sediments of the South China Sea. Microbial Ecology, 58(3): 582–590. doi: 10.1007/s00248-009-9506-z Zhou Mingyang, Wang Guanglong, Li Dan, et al. 2013. Diversity of both the cultivable protease-producing bacteria and bacterial extracellular proteases in the coastal sediments of King George Island, Antarctica. PLoS One, 8(11): e79668. doi: 10.1371/journal.pone.0079668 -

 点击查看大图
点击查看大图
计量
- 文章访问数: 818
- HTML全文浏览量: 257
- 被引次数: 0



 下载:
下载: