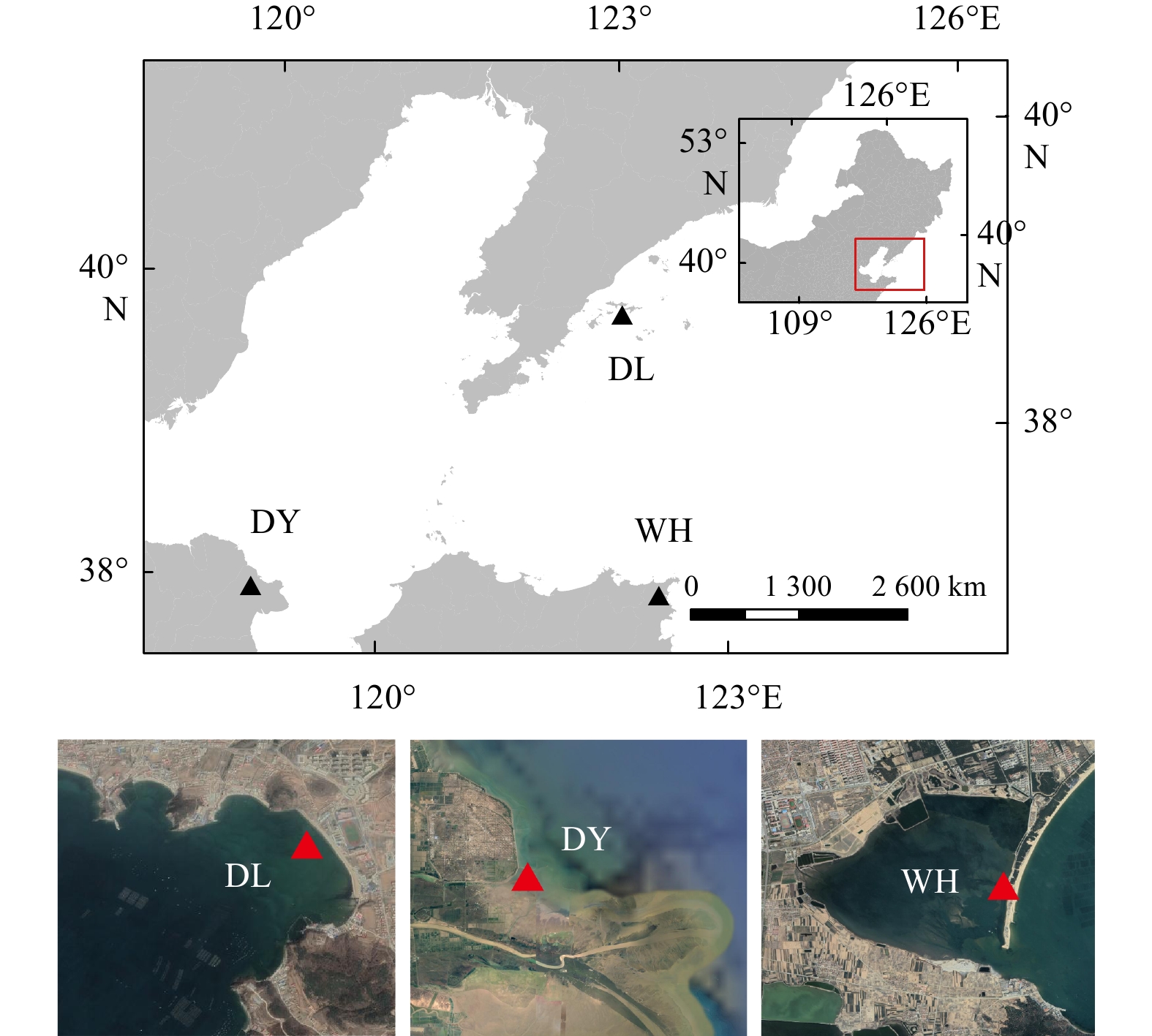
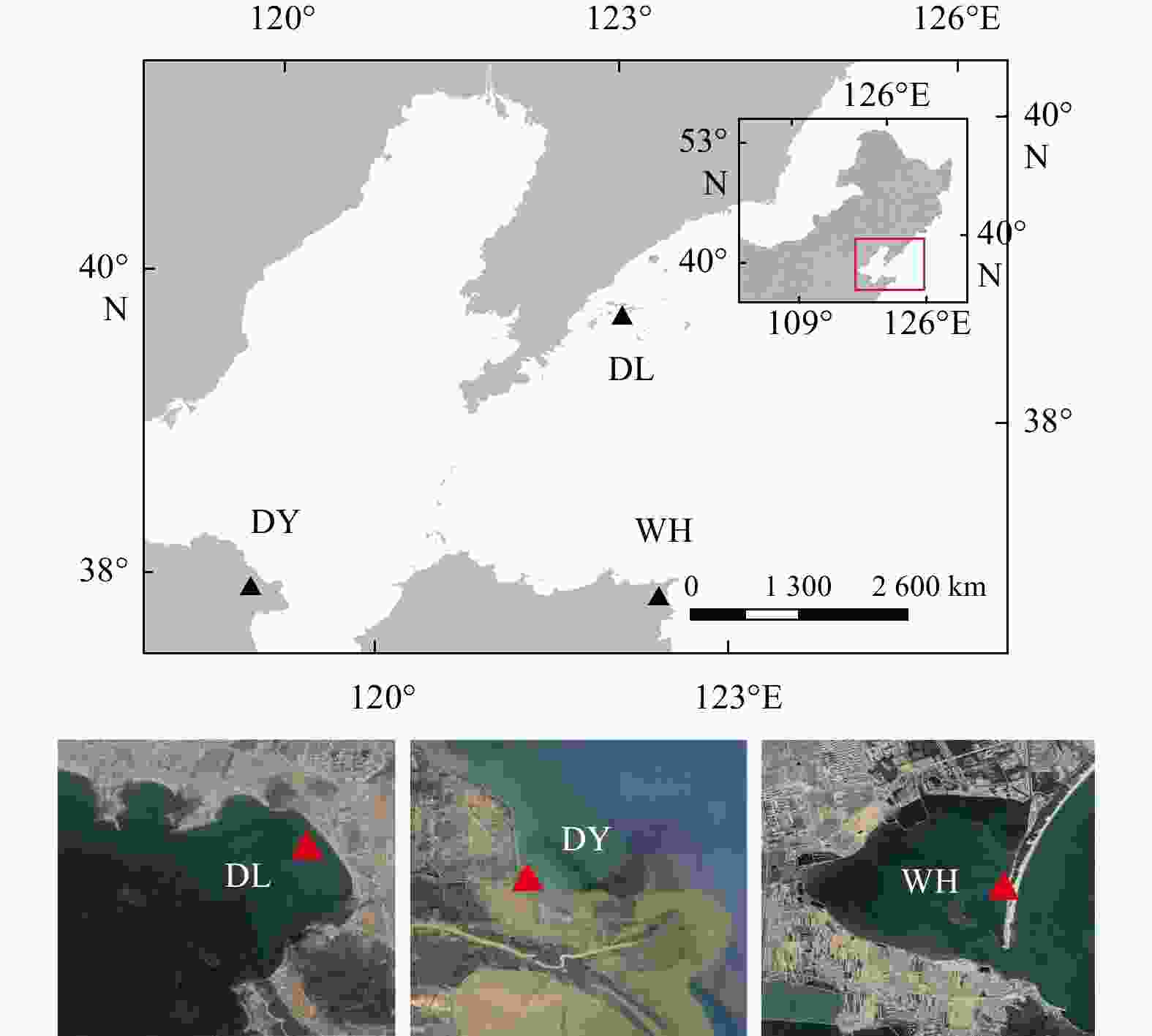
Molecular diversity and biogeography of benthic microeukaryotes in temperate seagrass (Zostera japonica) systems of northern China
-
Abstract: The productivity and health of seagrass depend on the combined inputs of nutrients from the water and sediments in which they grow and the microbiota with which they live intimately. However, little is known about the composition and diversity pattern of single-celled benthic eukaryotes in seagrass meadows. Here, we investigated how the structure and diversity of the benthic microeukaryotic community vary with respect to season, location, and seagrass colonization, by applying 18S rRNA gene amplicon sequencing for 96 surface sediment samples that were collected from three different seagrass habitats through four seasons. We found that benthic microeukaryotic communities associated with seagrass Zostera japonica exhibited remarkable spatial and seasonal variations, as well as differences between vegetated and unvegetated sediments. Diatoms and dinoflagellates predominated in the benthic microeukaryotic communities, but they were inversely correlated and displaced each other as the dominant microbial group in different seasons or habitats. Mucoromycota was more prevalent in vegetated sediments, whereas Lobulomycetales and Chytridiales had higher proportions in unvegetated sites. Total organic carbon and total organic nitrogen were the most important environmental factors in driving the microeukaryotic assemblages and diversity. Our study expands the available knowledge on the biogeographic distribution patterns and niche preferences for benthic microeukaryotes in seagrass systems.
-
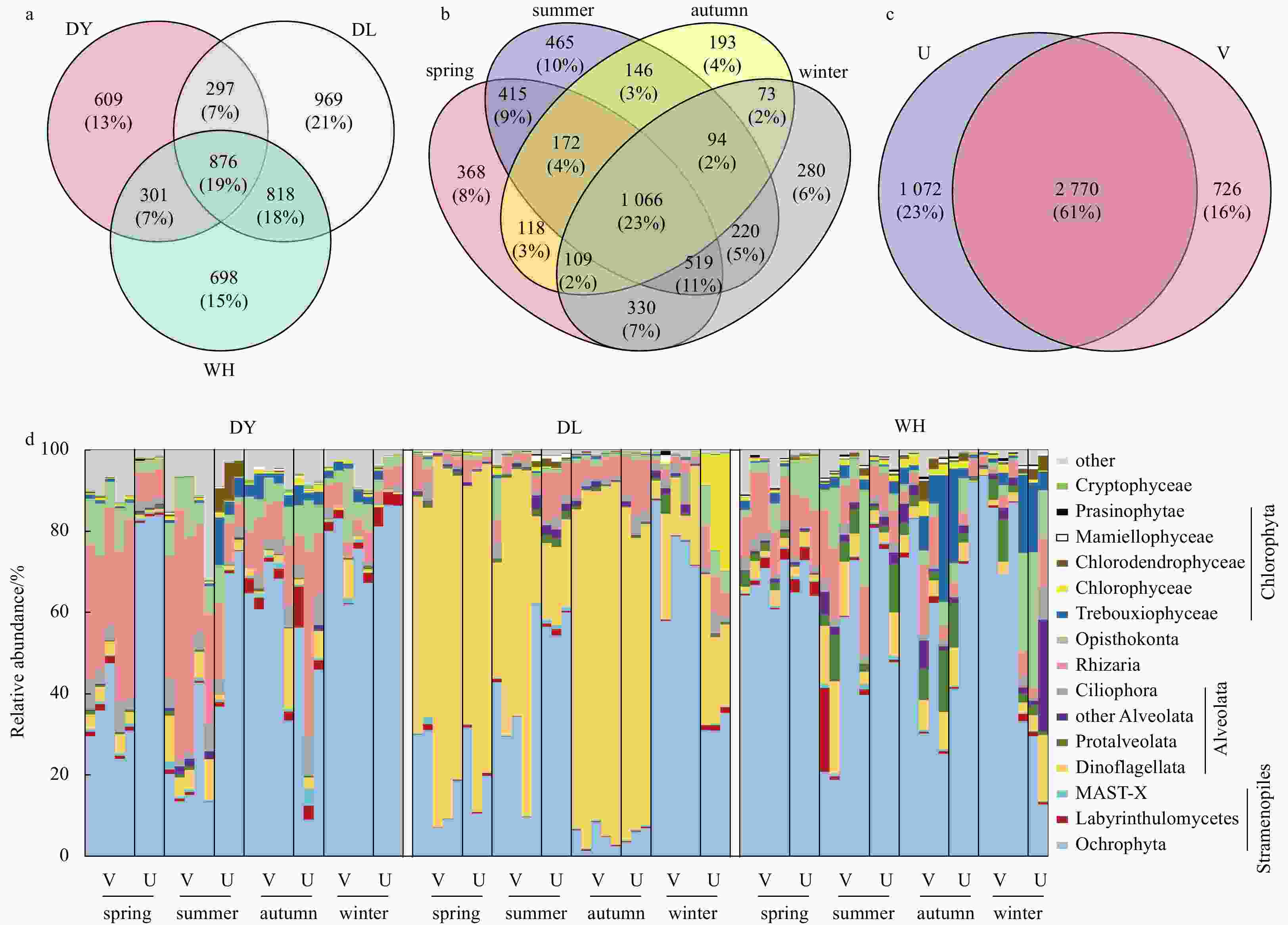
Figure 2. Venn diagrams showing the shared and unique OTU number in sediments among three sampling locations (a), four seasons (b), and seagrass vegetated (V) and unvegetated (U) samples (c). Taxonomic composition of seagrass Zostera japonica-associated microeukaryotic communities revealed by Miseq sequencing of 18S rRNA genes (d). The percentages in parentheses indicate the proportion of a given OTU number to the total. MAST: marine stramenopile.
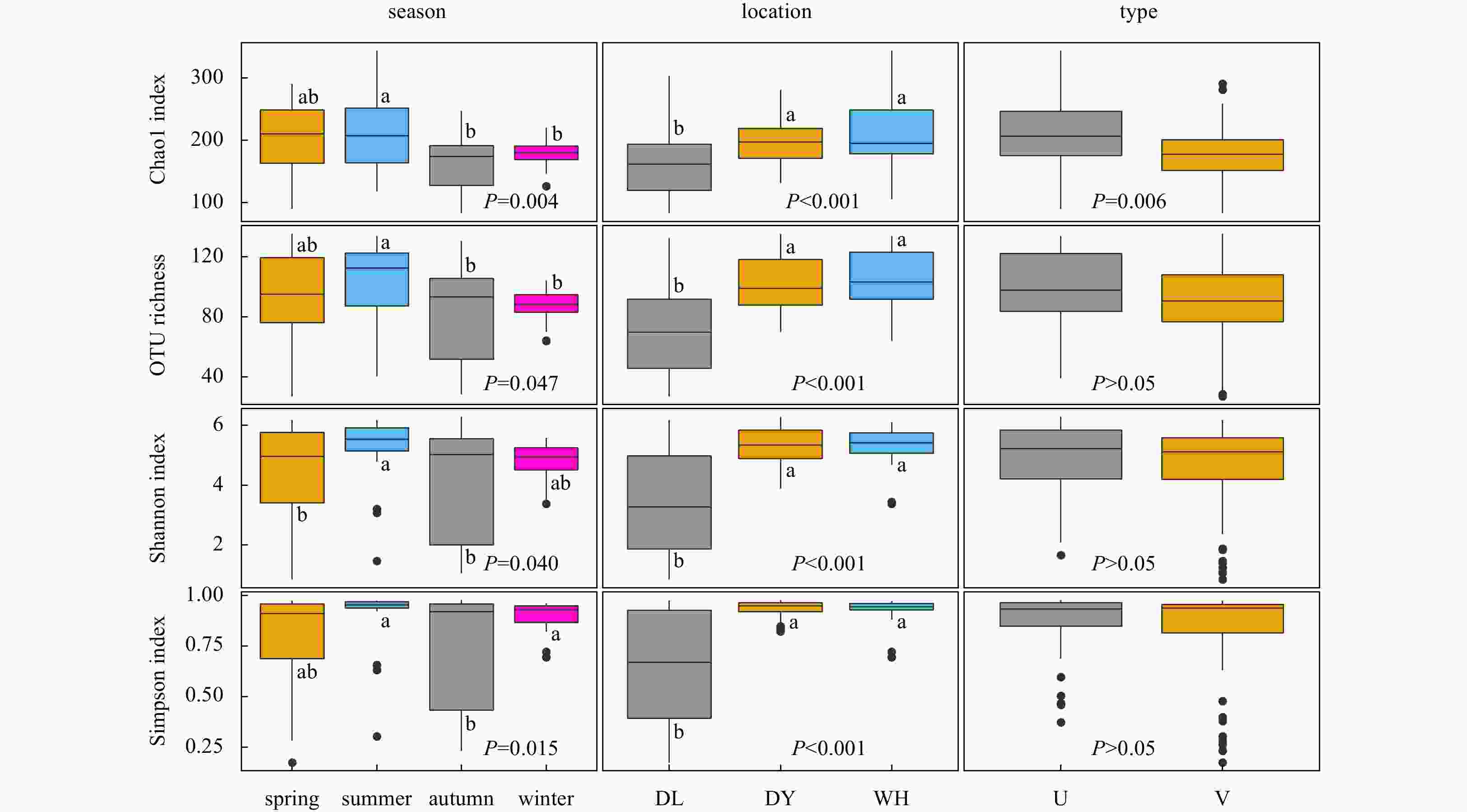
Figure 3. Comparisons of microeukaryotic α-diversity estimators of seagrass-vegetated (V) and unvegetated (U) sediments collected from Dongying (DY), Dalian (DL), and Weihai (WH) across four seasons. The P values are given for the comparison among seasons and locations using one-way ANOVA with least significance difference post hoc, while difference between vegetated and unvegetated was examined using t-test. Different letters above the box indicate significant differences among groups.
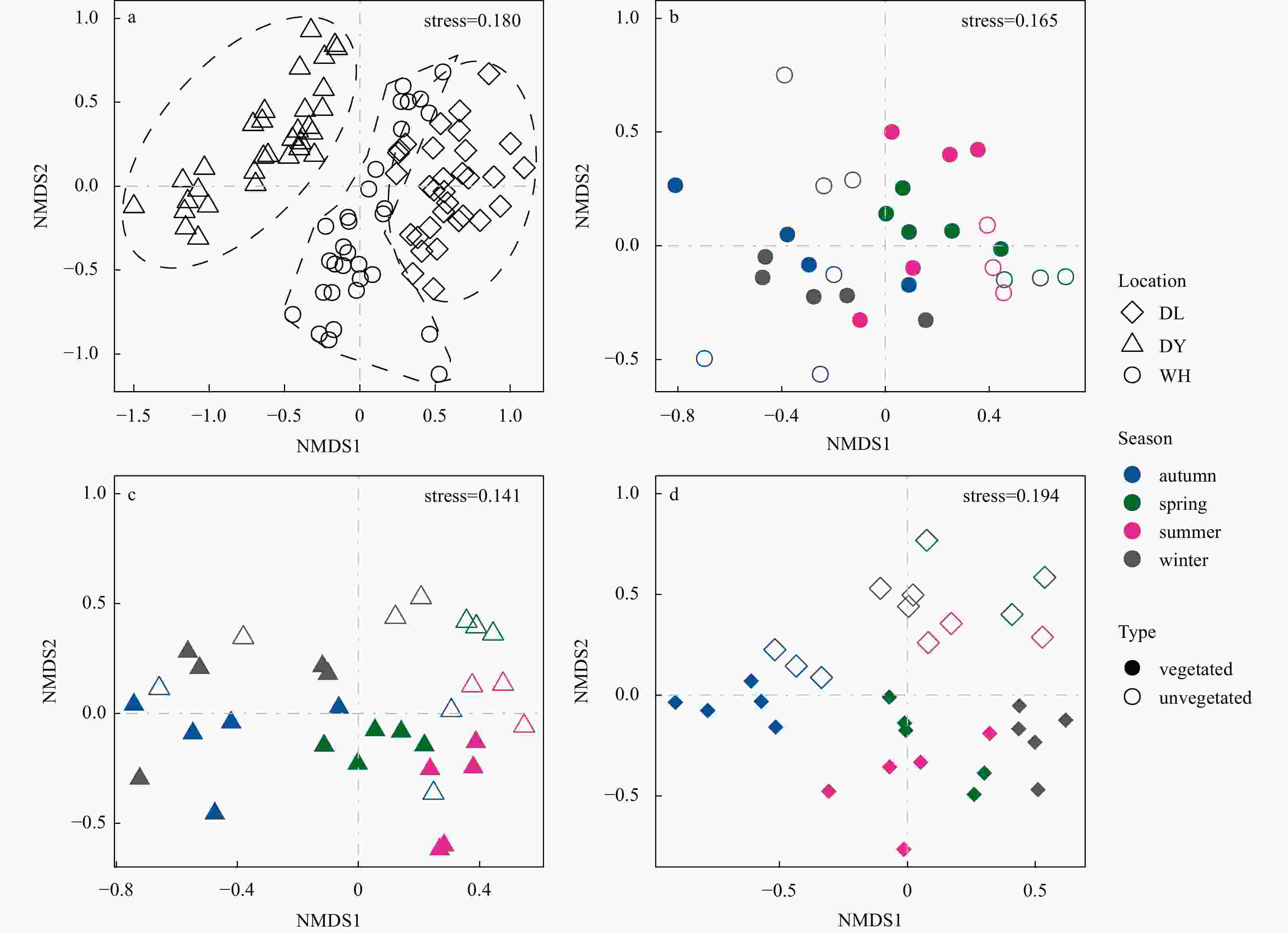
Figure 4. Plots of non-metric multidimensional scaling (NMDS) based on the Bray-Curtis distance, showing the variations of β-diversity of the microeukaryotic community from seagrass-vegetated (solid, V) and unvegetated (hollow, U) sediments in spatial scale (a), as well as seasonal patterns within each habitat: b in Weihai (WH); c in Dongying (DY), d in Dalian (DL). The shapes of points represent different location samples: DL, diamond; DY, triangle; and WH, round. And samples are colored by their corresponding seasons: spring, green; summer, pink; autumn, blue; and winter, black.
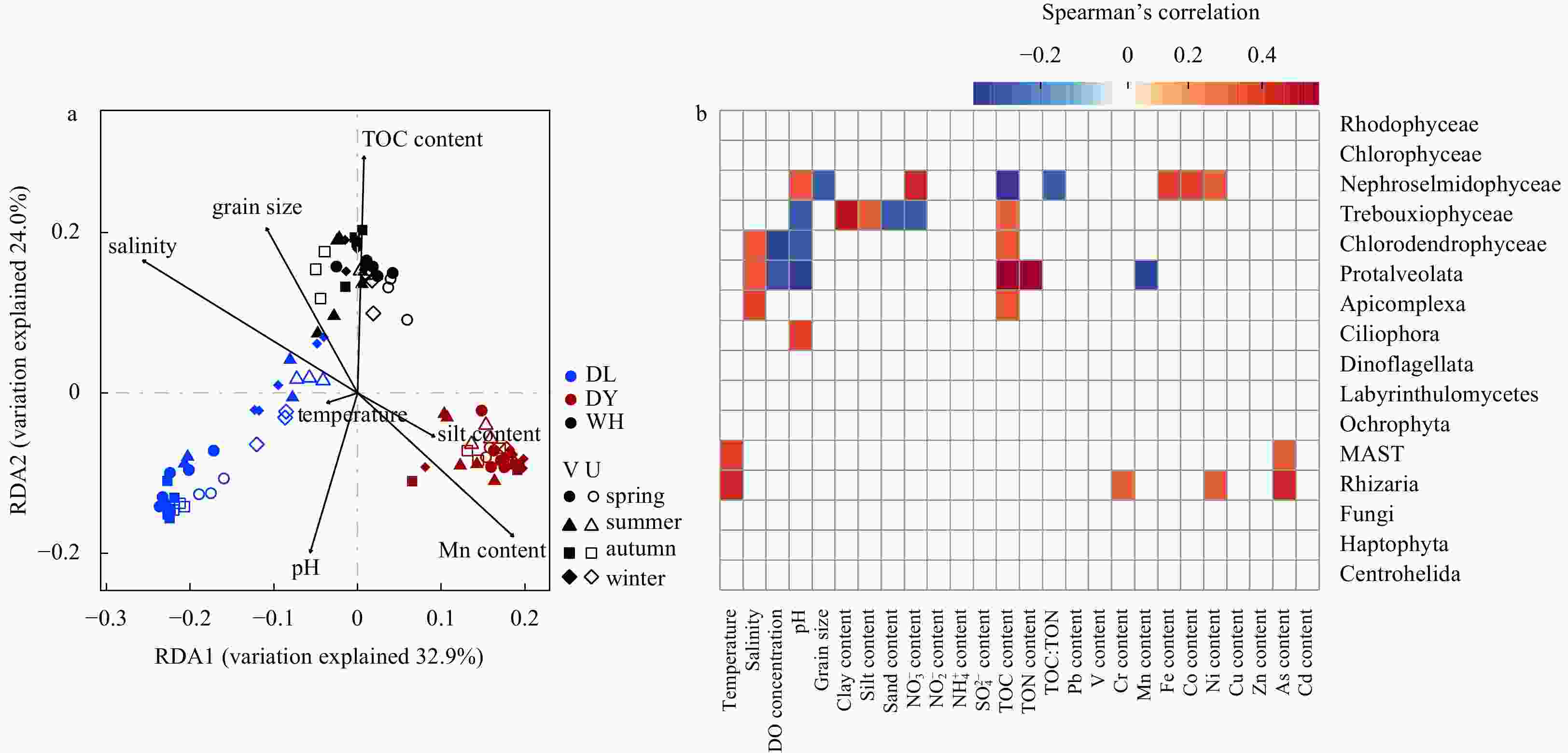
Figure 5. Redundancy analysis (RDA) ordination diagram (a) and heatmap of Spearman’s correlation (b) showing the relationships of geochemical variables with microeukaryotic community structure and relative abundances of major taxa, respectively. Letters in sample IDs mean the vegetated (V, solid) and unvegetated (U, hollow) samples, and shapes of points represent different season samples: spring, round; summer, triangle; autumn, square; and winter, diamond. Samples are colored by habitat types: DL, Dalian (sandy coast, blue); DY, Dongying (muddy coast, red); and WH, Weihai (lagoon, black). Only vectors for environmental variables with P-values of 0.05 or smaller are shown in a and b. The values of Spearman’s correlation coefficients are indicated according to the scale bar. MAST is the abbreviation of marine stramenopile; TOC:TON, the contents ratio between total organic carbon and organic nitrogen.
Table 1. ANOSIM testing the differences of benthic microeukaryotic communities between the two sediment types and four seasons within three seagrass beds, based on Bray-Curtis distance
Groups DL DY WH R P R P R P Global test 0.553 0.001 0.445 0.001 0.381 0.001 Spring vs. Summer 0.185 0.040 0.301 0.008 0.199 0.039 Spring vs. Autumn 0.568 0.001 0.451 0.001 0.658 0.001 Spring vs. Winter 0.486 0.002 0.491 0.002 0.455 0.001 Summer vs. Autumn 0.751 0.001 0.547 0.001 0.440 0.002 Summer vs. Winter 0.316 0.011 0.693 0.001 0.458 0.002 Autumn vs. Winter 0.901 0.001 0.248 0.015 0.081 0.125 Vegetated vs. Unvegetated 0.195 0.001 0.235 0.005 0.221 0.002 Note: Significant P-values (≤0.05) are highlighted in bold. Table 2. Spearman rank correlations between microeukaryotic α-diversity estimators and environment variables
Variable OTU richness Chao1 Simpson Shannon $\rho $ P $\rho $ P $\rho $ P $\rho $ P Temperature 0.35 0.001 0.23 0.029 0.33 0.001 0.37 <0.001 DO
concentration−0.22 0.034 −0.21 0.046 – – −0.21 0.043 pH −0.23 0.028 – – −0.21 0.044 −0.22 0.032 Grain size – – 0.22 0.034 – – – – ${{\rm {NH}}_4^+} $-N content −0.33 0.001 −0.34 0.001 −0.23 0.026 −0.28 0.007 ${{\rm {SO}}_4^{2-}} $ content – – – – 0.21 0.044 0.26 0.012 TOC content 0.29 0.005 0.22 0.031 0.28 0.005 0.31 0.003 TON content 0.33 0.001 0.26 0.010 0.33 0.001 0.36 <0.001 V content – – – – 0.21 0.047 – – Cr content – – – – 0.21 0.045 0.21 0.038 Note: Only the significant correlations (P≤0.05) are shown. OTU is the abbreviation of operational taxonomic unit. $\rho $ represents Spearman rank correlation coefficient; –, no data. -
Arasamuthu A, Mathews G, Patterson Edward J K. 2017. Spatial differences in bacterial and water quality parameters in seagrass meadows of Tuticorin Coast, Gulf of Mannar, southeastern India. Journal of Aquatic Biology & Fisheries, 5: 1–10 Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nature Reviews Microbiology, 5(10): 782–791. doi: 10.1038/nrmicro1747 Behera P, Mohapatra M, Kim J Y, et al. 2019. Spatial and temporal heterogeneity in the structure and function of sediment bacterial communities of a tropical mangrove forest. Environmental Science and Pollution Research, 26(4): 3893–3908. doi: 10.1007/s11356-018-3927-5 Bengtsson M M, Bühler A, Brauer A, et al. 2017. Eelgrass leaf surface microbiomes are locally variable and highly correlated with epibiotic eukaryotes. Frontiers in Microbiology, 8: 1312. doi: 10.3389/fmicb.2017.01312 Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57(1): 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x Caporaso J G, Kuczynski J, Stombaugh J, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7(5): 335–336. doi: 10.1038/nmeth.f.303 Clarke K R. 1993. Non-parametric multivariate analyses of changes in community structure. Australian Journal of Ecology, 18(1): 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x Cox T E, Cebrian J, Tabor M, et al. 2020. Do diatoms dominate benthic production in shallow systems? A case study from a mixed seagrass bed. Limnology and Oceanography Letters, 5(6): 425–434. doi: 10.1002/lol2.10167 Crump B C, Wojahn J M, Tomas F, et al. 2018. Metatranscriptomics and amplicon sequencing reveal mutualisms in seagrass microbiomes. Frontiers in Microbiology, 9: 388. doi: 10.3389/fmicb.2018.00388 Den Hartog C. 1989. Distribution of Plasmodiophora bicaudata, a parasitic fungus on small Zostera species. Diseases of Aquatic Organisms, 6: 227–229. doi: 10.3354/dao006227 Devereux R. 2005. Seagrass rhizosphere microbial communities. In: Kristensen E, Haese R R, Kostka J E, eds. Interactions Between Macro- and Microorganisms in Marine Sediments. Washington: American Geophysical Union, 199–216 Duarte C M, Holmer M, Marbà N. 2005. Plant-microbe interactions in seagrass meadows. In: Kristensen E, Haese R R, and Kostka J E, eds. Interactions Between Macro- and Microorganisms in Marine Sediments. Washington: American Geophysical Union, 31–60 Edgar R C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26(19): 2460–2461. doi: 10.1093/bioinformatics/btq461 Elwood H J, Olsen G J, Sogin M L. 1985. The small-subunit ribosomal RNA gene sequences from the hypotrichous ciliates Oxytricha nova and Stylonychia pustulata. Molecular Biology and Evolution, 2(5): 399–410. doi: 10.1093/oxfordjournals.molbev.a040362 Ettinger C L, Williams S L, Abbott J M, et al. 2017. Microbiome succession during ammonification in eelgrass bed sediments. PeerJ, 5: e3674. doi: 10.7717/peerj.3674 Frederiksen M S, Glud R N. 2006. Oxygen dynamics in the rhizosphere of Zostera marina: a two-dimensional planar optode study. Limnology and Oceanography, 51(2): 1072–1083. doi: 10.4319/lo.2006.51.2.1072 Frenken T, Alacid E, Berger S A, et al. 2017. Integrating chytrid fungal parasites into plankton ecology: research gaps and needs. Environmental Microbiology, 19(10): 3802–3822. doi: 10.1111/1462-2920.13827 Glücksman E, Bell T, Griffiths R I, et al. 2010. Closely related protist strains have different grazing impacts on natural bacterial communities. Environmental Microbiology, 12(12): 3105–3113. doi: 10.1111/j.1462-2920.2010.02283.x Gnavi G, Ercole E, Panno L, et al. 2014. Dothideomycetes and Leotiomycetes sterile mycelia isolated from the Italian seagrass Posidonia oceanica based on rDNA data. SpringerPlus, 3(1): 508. doi: 10.1186/2193-1801-3-508 Gong Jun, Dong Jun, Liu Xihan, et al. 2013. Extremely high copy numbers and polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist, 164(3): 369–379. doi: 10.1016/j.protis.2012.11.006 Gong Jun, Shi Fei, Ma Bin, et al. 2015. Depth shapes α- and β-diversities of microbial eukaryotes in surficial sediments of coastal ecosystems. Environmental Microbiology, 17(10): 3722–3737. doi: 10.1111/1462-2920.12763 Hansen J W, Udy J W, Perry C J, et al. 2000. Effect of the seagrass Zostera capricorni on sediment microbial processes. Marine Ecology Progress Series, 199: 83–96. doi: 10.3354/meps199083 Hanson C A, Fuhrman J A, Horner-Devine M C, et al. 2012. Beyond biogeographic patterns: processes shaping the microbial landscape. Nature Reviews Microbiology, 10(7): 497–506. doi: 10.1038/nrmicro2795 Hurtado-McCormick V, Kahlke T, Petrou K, et al. 2019. Regional and microenvironmental scale characterization of the Zostera muelleri seagrass microbiome. Frontiers in Microbiology, 10: 1011. doi: 10.3389/fmicb.2019.01011 Ikenaga M, Guevara R, Dean A L, et al. 2010. Changes in community structure of sediment bacteria along the Florida coastal everglades marsh-mangrove-seagrass salinity gradient. Microbial Ecology, 59(2): 284–295. doi: 10.1007/s00248-009-9572-2 James J B, Sherman T D, Devereux R. 2006. Analysis of bacterial communities in seagrass bed sediments by double-gradient denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA genes. Microbial Ecology, 52(4): 655–661. doi: 10.1007/s00248-006-9075-3 Jensen S I, Kühl M, Glud R N, et al. 2005. Oxic microzones and radial oxygen loss from roots of Zostera marina. Marine Ecology Progress Series, 293: 49–58. doi: 10.3354/meps293049 Kosa G, Zimmermann B, Kohler A, et al. 2018. High-throughput screening of Mucoromycota fungi for production of low- and high-value lipids. Biotechnology for Biofuels, 11(1): 66. doi: 10.1186/s13068-018-1070-7 Krabberød A K, Bjorbækmo M F M, Shalchian-Tabrizi K, et al. 2017. Exploring the oceanic microeukaryotic interactome with metaomics approaches. Aquatic Microbial Ecology, 79(1): 1–12. doi: 10.3354/ame01811 Küçük A, Ergül H A. 2011. Seasonal variations of microplankton composition in İzmit Bay (Sea of Marmara). Journal of the Black Sea/Mediterranean Environment, 17(3): 216–222 Lallias D, Hiddink J G, Fonseca V G, et al. 2015. Environmental metabarcoding reveals heterogeneous drivers of microbial eukaryote diversity in contrasting estuarine ecosystems. The ISME Journal, 9(5): 1208–1221. doi: 10.1038/ismej.2014.213 Lear G, Bellamy J, Case B S, et al. 2014. Fine-scale spatial patterns in bacterial community composition and function within freshwater ponds. The ISME Journal, 8(8): 1715–1726. doi: 10.1038/ismej.2014.21 Lehnen N, Marchant H K, Schwedt A, et al. 2016. High rates of microbial dinitrogen fixation and sulfate reduction associated with the Mediterranean seagrass Posidonia oceanica. Systematic and Applied Microbiology, 39(7): 476–483. doi: 10.1016/j.syapm.2016.08.004 Lin Haiying, Sun Tao, Xue Sufeng, et al. 2016. Heavy metal spatial variation, bioaccumulation, and risk assessment of Zostera japonica habitat in the Yellow River Estuary, China. Science of the Total Environment, 541: 435–443. doi: 10.1016/j.scitotenv.2015.09.050 Lin Xianbiao, Zheng Pengfei, Zou Songbao, et al. 2021. Seagrass (Zostera marina) promotes nitrification potential and selects specific ammonia oxidizers in coastal sediments. Journal of Soils and Sediments, 21(10): 3259–3273. doi: 10.1007/s11368-021-02951-w Ling Juan, Zhang Yanying, Wu Meilin, et al. 2015. Fungal community successions in rhizosphere sediment of seagrasses Enhalus acoroides under PAHs stress. International Journal of Molecular Sciences, 16(6): 14039–14055. doi: 10.3390/ijms160614039 Liu Lemian, Wang Shanshan, Chen Jianfeng. 2020. Hysteretic response of microbial eukaryotic communities to gradually decreased nutrient concentrations in eutrophic water. Microbial Ecology, 79(4): 815–822. doi: 10.1007/s00248-019-01457-w Magdouli S, Yan S, Tyagi R D, et al. 2014. Heterotrophic microorganisms: a promising source for biodiesel production. Critical Reviews in Environmental Science and Technology, 44(4): 416–453. doi: 10.1080/10643389.2012.728523 Malviya S, Scalco E, Audic S, et al. 2016. Insights into global diatom distribution and diversity in the world’s ocean. Proceedings of the National Academy of Sciences of the United States of America, 113(11): E1516–E1525. doi: 10.1073/pnas.1509523113 Marquardt M, Vader A, Stübner E I, et al. 2016. Strong seasonality of marine microbial eukaryotes in a high-Arctic fjord (Isfjorden, in West Spitsbergen, Norway). Applied and Environmental Microbiology, 82(6): 1868–1880. doi: 10.1128/AEM.03208-15 Martiny J B H, Bohannan B J M, Brown J H, et al. 2006. Microbial biogeography: putting microorganisms on the map. Nature Reviews Microbiology, 4(2): 102–112. doi: 10.1038/nrmicro1341 Maslennikova S, Larina N, Larin S. 2012. The effect of sediment grain size on heavy metal content. Lakes Reservoirs and Ponds, 6(1): 43–54 Massana R, Gobet A, Audic S, et al. 2015. Marine protist diversity in European coastal waters and sediments as revealed by high-throughput sequencing. Environmental Microbiology, 17(10): 4035–4049. doi: 10.1111/1462-2920.12955 McMurdie P J, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Computational Biology, 10(4): e1003531. doi: 10.1371/journal.pcbi.1003531 Medina-Pons F J, Terrados J, López-López A, et al. 2009. Evaluation of the 18S rRNA clone library approach to study the diversity of the macroeukaryotic leaf-epiphytic community of the seagrass Posidonia oceanica (L.) Delile. Marine Biology, 156(9): 1963–1976. doi: 10.1007/s00227-009-1221-2 Nelson D M, Tréguer P, Brzezinski M A, et al. 1995. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochemical Cycles, 9(3): 359–372. doi: 10.1029/95GB01070 Newell S Y. 1981. Fungi and bacteria in or on leaves of eelgrass (Zostera marina L.) from Chesapeake Bay. Applied and Environmental Microbiology, 41(5): 1219–1224. doi: 10.1128/aem.41.5.1219-1224.1981 Oberbeckmann S, Loeder M G J, Gerdts G, et al. 2014. Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters. FEMS Microbiology Ecology, 90(2): 478–492. doi: 10.1111/1574-6941.12409 Onishi Y, Mohri Y, Tuji A, et al. 2014. The seagrass Zostera marina harbors growth-inhibiting bacteria against the toxic dinoflagellate Alexandrium tamarense. Fisheries Science, 80(2): 353–362. doi: 10.1007/s12562-013-0688-4 Orsi W, Song Y C, Hallam S, et al. 2012. Effect of oxygen minimum zone formation on communities of marine protists. The ISME Journal, 6(8): 1586–1601. doi: 10.1038/ismej.2012.7 Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics, 20(2): 289–290. doi: 10.1093/bioinformatics/btg412 Petersen L E, Marner M, Labes A, et al. 2019. Rapid metabolome and bioactivity profiling of fungi associated with the leaf and rhizosphere of the Baltic seagrass Zostera marina. Marine Drugs, 17(7): 419. doi: 10.3390/md17070419 Robinson M D, McCarthy D J, Smyth G K. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1): 139–140. doi: 10.1093/bioinformatics/btp616 Sakayaroj J, Preedanon S, Supaphon O, et al. 2010. Phylogenetic diversity of endophyte assemblages associated with the tropical seagrass Enhalus acoroides in Thailand. Fungal Diversity, 42(1): 27–45. doi: 10.1007/s13225-009-0013-9 Schloss P D, Westcott S L, Ryabin T, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75(23): 7537–7541. doi: 10.1128/AEM.01541-09 Sherr E B, Sherr B F. 2002. Significance of predation by protists in aquatic microbial food webs. Antonie van Leeuwenhoek, 81(1): 293–308. doi: 10.1023/A:1020591307260 Short F T, Polidoro B, Livingstone S R, et al. 2011. Extinction risk assessment of the world’s seagrass species. Biological Conservation, 144(7): 1961–1971. doi: 10.1016/j.biocon.2011.04.010 Smayda T J, Trainer V L. 2010. Dinoflagellate blooms in upwelling systems: Seeding, variability, and contrasts with diatom bloom behaviour. Progress in Oceanography, 85(1–2): 92–107, Smith A C, Kostka J E, Devereux R, et al. 2004. Seasonal composition and activity of sulfate-reducing prokaryotic communities in seagrass bed sediments. Aquatic Microbial Ecology, 37(2): 183–195. doi: 10.3354/ame037183 Sun Feifei, Zhang Xiaoli, Zhang Qianqian, et al. 2015. Seagrass (Zostera marina) colonization promotes the accumulation of diazotrophic bacteria and alters the relative abundances of specific bacterial lineages involved in benthic carbon and sulfur cycling. Applied and Environmental Microbiology, 81(19): 6901–6914. doi: 10.1128/aem.01382-15 Trevizan Segovia B, Sanders-Smith R, Adamczyk E M, et al. 2021. Microeukaryotic communities associated with the seagrass Zostera marina are spatially structured. Journal of Eukaryotic Microbiology, 68: e12827. doi: 10.1111/jeu.12827 Tuchman N C, Schollett M A, Rier S T, et al. 2006. Differential heterotrophic utilization of organic compounds by diatoms and bacteria under light and dark conditions. Hydrobiologia, 561(1): 167–177. doi: 10.1007/s10750-005-1612-1614 Ugarelli K, Chakrabarti S, Laas P, et al. 2017. The seagrass holobiont and its microbiome. Microorganisms, 5(4): 81. doi: 10.3390/microorganisms5040081 Van den Wyngaert S, Rojas-Jimenez K, Seto K, et al. 2018. Diversity and hidden host specificity of chytrids infecting colonial volvocacean algae. Journal of Eukaryotic Microbiology, 65(6): 870–881. doi: 10.1111/jeu.12632 Virta L, Soininen J, Norkko A. 2020. Stable seasonal and annual alpha diversity of benthic diatom communities despite changing community composition. Frontiers in Marine Science, 7: 88. doi: 10.3389/fmars.2020.00088 Wainwright B J, Zahn G L, Arlyza I S, et al. 2018. Seagrass-associated fungal communities follow Wallace’s line, but host genotype does not structure fungal community. Journal of Biogeography, 45(4): 762–770. doi: 10.1111/jbi.13168 Walker A K, Campbell J. 2009. First records of the seagrass parasite Plasmodiophora diplantherae from the northcentral Gulf of Mexico. Gulf and Caribbean Research, 21(1): 63–65. doi: 10.18785/gcr.2101.07 Wang Yongming, Liu Lemian, Chen Huihuang, et al. 2015. Spatiotemporal dynamics and determinants of planktonic bacterial and microeukaryotic communities in a Chinese subtropical river. Applied Microbiology and Biotechnology, 99(21): 9255–9266. doi: 10.1007/s00253-015-6773-0 Wu Pengfei, Li Dongxu, Kong Lingfen, et al. 2020. The diversity and biogeography of microeukaryotes in the euphotic zone of the northwestern Pacific Ocean. Science of the Total Environment, 698: 134289. doi: 10.1016/j.scitotenv.2019.134289 Xiao Wupeng, Liu Xin, Irwin A J, et al. 2018. Warming and eutrophication combine to restructure diatoms and dinoflagellates. Water Research, 128: 206–216. doi: 10.1016/j.watres.2017.10.051 Yamada M, Otsubo M, Kodama M, et al. 2014. Species composition of Skeletonema (Bacillariophyceae) in planktonic and resting-stage cells in Osaka and Tokyo Bays. Plankton and Benthos Research, 9(3): 168–175. doi: 10.3800/pbr.9.168 Zettler L A A, Gómez F, Zettler E, et al. 2002. Eukaryotic diversity in Spain’s River of Fire. Nature, 417(6885): 137. doi: 10.1038/417137a Zhang Wenjing, Pan Yongbo, Yang Jun, et al. 2018. The diversity and biogeography of abundant and rare intertidal marine microeukaryotes explained by environment and dispersal limitation. Environmental Microbiology, 20(2): 462–476. doi: 10.1111/1462-2920.13916 Zheng Pengfei, Wang Chuantao, Zhang Xiaoli, et al. 2019. Community structure and abundance of archaea in a Zostera marina meadow: a comparison between seagrass-colonized and bare sediment sites. Archaea, 2019: 5108012. doi: 10.1155/2019/5108012 Zhu Ping, Wang Yaping, Shi Tiantian, et al. 2018. Genetic diversity of benthic microbial eukaryotes in response to spatial heterogeneity of sediment geochemistry in a mangrove ecosystem. Estuaries and Coasts, 41(3): 751–764. doi: 10.1007/s12237-017-0317-z -




 下载:
下载: